04 Jan 2014
Editor’s note: This is a previously published post of mine from a couple of years ago (!). I always thought about turning it into a paper. The interesting idea (I think) is how the causal model matters for whether the lasso or the marginal regression approach works better. Also check it out, the Leekasso is part of the SuperLearner package.
One incredibly popular tool for the analysis of high-dimensional data is the lasso. The lasso is commonly used in cases when you have many more predictors than independent samples (the n « p) problem. It is also often used in the context of prediction.
Suppose you have an outcome Y and several predictors X1,…,XM, the lasso fits a model:
Y = B0 + B1 X1 + B2 X2 + … + BM XM + E
subject to a constraint on the sum of the absolute value of the B coefficients. The result is that: (1) some of the coefficients get set to zero, and those variables drop out of the model, (2) other coefficients are “shrunk” toward zero. Dropping some variables is good because there are a lot of potentially unimportant variables. Shrinking coefficients may be good, since the big coefficients might be just the ones that were really big by random chance (this is related to Andrew Gelman’s type M errors).
I work in genomics, where n«p problems come up all the time. Whenever I use the lasso or when I read papers where the lasso is used for prediction, I always think: “How does this compare to just using the top 10 most significant predictors?” I have asked this out loud enough that some people around here started calling it the “Leekasso” to poke fun at me. So I’m going to call it that in a thinly veiled attempt to avoid Stigler’s law of eponymy (actually Rafa points out that using this name is a perfect example of this law, since this feature selection approach has been proposed before at least once).
Here is how the Leekasso works. You fit each of the models:
Y = B0 + BkXk + E
take the 10 variables with the smallest p-values from testing the Bk coefficients, then fit a linear model with just those 10 coefficients. You never use 9 or 11, the Leekasso is always 10.
For fun I did an experiment to compare the accuracy of the Leekasso and the Lasso.
Here is the setup:
- I simulated 500 variables and 100 samples for each study, each N(0,1)
- I created an outcome that was 0 for the first 50 samples, 1 for the last 50
- I set a certain number of variables (between 5 and 50) to be associated with the outcome using the model Xi = b0i + b1iY + e (this is an important choice, more later in the post)
- I tried different levels of signal to the truly predictive features
- I generated two data sets (training and test) from the exact same model for each scenario
- I fit the Lasso using the lars package, choosing the shrinkage parameter as the value that minimized the cross-validation MSE in the training set
- I fit the Leekasso and the Lasso on the training sets and evaluated accuracy on the test sets.
The R code for this analysis is available here and the resulting data is here.
The results show that for all configurations, using the top 10 has a higher out of sample prediction accuracy than the lasso. A larger version of the plot is here.
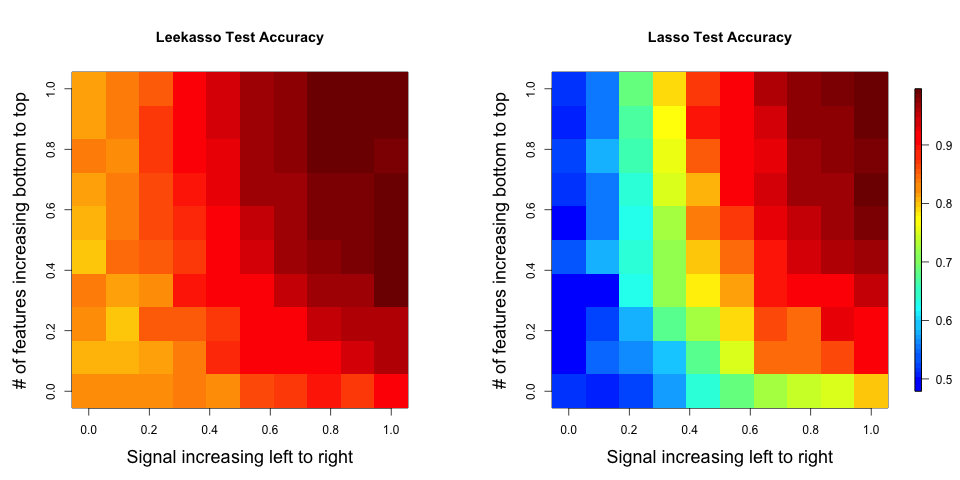
Interestingly, this is true even when there are fewer than 10 real features in the data or when there are many more than 10 real features ((remember the Leekasso always picks 10).
Some thoughts on this analysis:
- This is only test-set prediction accuracy, it says nothing about selecting the “right” features for prediction.
- The Leekasso took about 0.03 seconds to fit and test per data set compared to about 5.61 seconds for the Lasso.
- The data generating model is the model underlying the top 10, so it isn’t surprising it has higher performance. Note that I simulated from the model: Xi = b0i + b1iY + e, this is the model commonly assumed in differential expression analysis (genomics) or voxel-wise analysis (fMRI). Alternatively I could have simulated from the model: Y = B0 + B1 X1 + B2 X2 + … + BM XM + E, where most of the coefficients are zero. In this case, the Lasso would outperform the top 10 (data not shown). This is a key, and possibly obvious, issue raised by this simulation. When doing prediction differences in the true “causal” model matter a lot. So if we believe the “top 10 model” holds in many high-dimensional settings, then it may be the case that regularization approaches don’t work well for prediction and vice versa.
- I think what may be happening is that the Lasso is overshrinking the parameter estimates, in other words, you give up too much bias for a gain in variance. Alan Dabney and John Storey have a really nice paper discussing shrinkage in the context of genomic prediction that I think is related.
5.
03 Jan 2014
Recently, the U.S. Supreme Court heard arguments in the cases EPA v. EME Homer City Generation and _American Lung Association v EME Homer City Generation. _SCOTUSblog has a nice summary of the legal arguments, for the law buffs out there.
The basic problem is that the way air pollution is regulated, the EPA and state and local agencies monitor the air pollution in each state. When the levels of pollution are above the national ambient air quality standards at the monitors in that state, the state is considered in “non-attainment” (i.e. they have not attained the standard). Otherwise, they are in attainment.
But what if your state doesn’t actually generate any pollution, but there’s all this pollution blowing in from another state? Pollution knows no boundaries and in that case, the monitors in your state will be in non-attainment, and it isn’t even your fault! The Clean Air Act has something called the “good neighbor” policy that was designed to address this issue. From SCOTUSblog:
One of the obligations that states have, in drafting implementation plans [to reduce pollution], is imposed by what is called the “good neighbor” policy. It dates from 1963, in a more elemental form, but its most fully developed form requires each state to include in its plan the measures necessary to prevent the migration of their polluted air to their neighbors, if that would keep the neighbors from meeting EPA’s quality standards.
The problem is that if you live in a state like Maryland, your air pollution is coming from a bunch of states (Pennsylvania, Ohio, etc.). So who do you blame? Well, the logical thing would be to say that if Pennsylvania contributes to 90% of Maryland’s interstate air pollution and Ohio contributes 10%, then Pennsylvania should get 90% of the blame and Ohio 10%. But it’s not so easy because air pollution doesn’t have any special identifiers on it to indicate what state it came from. This is the source apportionment problem in air pollution and it involves trying to back-calculate where a given amount of pollution came from (or what was its source). It’s not an easy problem.
EPA realized the unfairness here and devised the State Air Pollution Rule, also known as the “Transport Rule”. From SCOTUSblog:
What the Transport Rule sought to do is to set up a regime to limit cross-border movement of emissions of nitrogen oxides and sulfur dioxide. Those substances, sent out from coal-fired power plants and other sources, get transformed into ozone and “fine particular matter” (basically, soot), and both are harmful to human health, contributing to asthma and heart attacks. They also damage natural terrain such as forests, destroy farm crops, can kill fish, and create hazes that reduce visibility.
Both of those pollutants are carried by the wind, and they can be transported very large distances — a phenomenon that is mostly noticed in the eastern states.
There are actually a few versions of this problem. One common one involves identifying the source of a particle (i.e. automobile, power plans, road dust) based on its chemical composition. The idea here is that at any given monitor, there are particles blowing in from all different types of sources and so the pollution you measure is a mixture of all these sources. Making some assumptions about chemical mass balance, there are ways to statistically separate out the contributions from individual sources based on a the chemical composition of the total mass measurement. If the particles that we measure, say, have a lot of ammonium ions and we know that particles generated by coal-burning power plants have a lot of ammonium ions, then we might infer that the particles came from a coal-burning power plant.
The key idea here is that different sources of particles have “chemical signatures” that can be used to separate out their various contributions. This is already a difficult problem, but at least here, we have some knowledge of the chemical makeup of various sources and can incorporate that knowledge into the statistical analysis.
In the problem at the Supreme Court, we’re not concerned with particles from various types of sources, but rather from different locations. But, for the most part, different states don’t have “chemical signatures” or tracer elements, so it’s hard to identify whether a given particle (or other pollutant) blowing in the wind came from Pennsylvania versus Ohio.
So what did EPA do? Well, instead of figuring out where the pollution came from, they decided that states would reduce emissions based on how much it would cost to control those emissions. The states objected because the cost of controlling emissions may well have nothing to do with how much pollution is actually being contributed downwind.
The legal question involves whether or not EPA has the authority to devise a regulatory plan based on costs as opposed to actual pollution contribution. I will let people who actually know the law address that question, but given the general difficulty of source apportionment, I’m not sure EPA could have come up with a much better plan.
30 Dec 2013
There is a lot of noise around the “R versus Contender X” for Data Science. I think the two main competitors right now that I hear about are Python and Julia. I’m not going to weigh into the debates because I go by the motto: “Why not just use something that works?”
R offers a lot of benefits if you are interested in statistical or predictive modeling. It is basically unrivaled in terms of the breadth of packages for applied statistics. But I think sometimes it isn’t obvious that R can handle some tasks that you used to have to do with other languages. This misconception is particularly common among people who regularly code in a different language and are moving to R. So I thought I’d point out a few cool things that R can do. Please add to the list in the comments if I’ve missed things that R can do people don’t expect.
- R can do regular expressions/text processing: Check out stringr, tm, and a large number of other natural language processing packages.
- R can get data out of a database: Check out RMySQL, RMongoDB, rhdf5, ROracle, MonetDB.R (via Anthony D.).
- R can process nasty data: Check out plyr, reshape2, Hmisc
- R can process images: EBImage is a good general purpose tool, but there are also packages for various file types like jpeg.
- R can handle different data formats: XML and RJSONIO handle two common types, but you can also read from Excel files with xlsx or handle pretty much every common data storage type (you’ll have to search R + data type) to find the package.
- R can interact with APIs: Check out RCurl and httr for general purpose software, or you could try some specific examples like twitteR. You can create an api from R code using yhat.
- R can build apps/interactive graphics: Some pretty cool things have already been built with shiny, rCharts interfaces with a ton of interactive graphics packages.
- R can create dynamic documents: Try out [There is a lot of noise around the “R versus Contender X” for Data Science. I think the two main competitors right now that I hear about are Python and Julia. I’m not going to weigh into the debates because I go by the motto: “Why not just use something that works?”
R offers a lot of benefits if you are interested in statistical or predictive modeling. It is basically unrivaled in terms of the breadth of packages for applied statistics. But I think sometimes it isn’t obvious that R can handle some tasks that you used to have to do with other languages. This misconception is particularly common among people who regularly code in a different language and are moving to R. So I thought I’d point out a few cool things that R can do. Please add to the list in the comments if I’ve missed things that R can do people don’t expect.
- R can do regular expressions/text processing: Check out stringr, tm, and a large number of other natural language processing packages.
- R can get data out of a database: Check out RMySQL, RMongoDB, rhdf5, ROracle, MonetDB.R (via Anthony D.).
- R can process nasty data: Check out plyr, reshape2, Hmisc
- R can process images: EBImage is a good general purpose tool, but there are also packages for various file types like jpeg.
- R can handle different data formats: XML and RJSONIO handle two common types, but you can also read from Excel files with xlsx or handle pretty much every common data storage type (you’ll have to search R + data type) to find the package.
- R can interact with APIs: Check out RCurl and httr for general purpose software, or you could try some specific examples like twitteR. You can create an api from R code using yhat.
- R can build apps/interactive graphics: Some pretty cool things have already been built with shiny, rCharts interfaces with a ton of interactive graphics packages.
- R can create dynamic documents: Try out](http://yihui.name/knitr/) or slidify.
- R can play with Hadoop: Check out the rhadoop wiki.
- R can create interactive teaching modules: You can do it in the console with swirl or on the web with Datamind.
- R interfaces very nicely with C if you need to be hardcore (also maybe? interfaces with Python): Rcpp, enough said. Also read the tutorial. I haven’t tried the rPython library, but it looks like a great idea.
20 Dec 2013
Editor’s Note: I made this list off the top of my head and have surely missed awesome things people have done this year. If you know of some, you should make your own list or add it to the comments! I have also avoided talking about stuff I worked on or that people here at Hopkins are doing because this post is supposed to be about other people’s awesome stuff. I wrote this post because a blog often feels like a place to complain, but we started Simply Stats as a place to be pumped up about the stuff people were doing with data.
- I emailed Hadley Wickham about some trouble we were having memory profiling. He wrote back immediately, then wrote an R package, then wrote this awesome guide. That guy is ridiculous.
- Jared Horvath wrote this incredibly well-written and compelling argument for the scientific system that has given us a wide range of discoveries.
- Yuwen Liu and colleagues wrote this really interesting paper on power for RNA-seq studies comparing biological replicates and sequencing depth. Shows pretty conclusively to go for more replicates (music to a statisticians ears!).
- Yoav Benjamini and Yotam Hechtlingler wrote an amazing discussion of the paper we wrote about the science-wise false discovery rate. It contributes new ideas about estimation/control in that context.
- Sherri Rose wrote a fascinating article about statistician’s role in big data. One thing I really liked was this line: “This may require implementing commonly used methods, developing a new method, or integrating techniques from other fields to answer our problem.” I really like the idea that integrating and applying standard methods in new and creative ways can be viewed as a statistical contribution.
- Karl Broman gave his now legendary talk (part1/part2) on statistical graphics that I think should be required viewing for anyone who will ever plot data on a Google Hangout with the Iowa State data viz crowd. They had some technical difficulties during the broadcast so Karl B. took it down. Join me in begging him to put it back up again despited the warts.
- Everything Thomas Lumley wrote on notstatschat, I follow that blog super closely. I love this scrunchable poster he pointed to and this post on Statins and the Causal Markov property.
- I wish I could take Joe Blitzstein’s data science class. Particularly check out the reading list, which I think is excellent.
- Lev Muchik, Sinan Aral, and Sean Taylor brought the randomized control trial to social influence bias on a massive scale. I love how RCT are finding their ways into the new, sexy areas.
- Genevera Allen taught a congressman about statistical brain mapping and holy crap he talked about it on the floor of the house.
- Lior Pachter starting mixing it up on his blog. I don’t necessarily agree with all of his posts but it is hard to deny the influence that his posts have had on real science. I definitely read it regularly.
- Marie Davidian, President of the ASA, has been on a tear this year, doing tons of cool stuff, including landing the big fish, Nate Silver, for JSM. Super impressive to watch the energy. I’m also really excited to see what Bin Yu works on this year as president of IMS.
- The Stats 2013 crowd has done a ridiculously good job of getting the word out about statistics this year. I keep seeing statistics pop up in places like the WSJ, which warms my heart.
- One way I judge a paper is by how angry/jealous I am that I didn’t think of or write that paper. This paper on the reproducibility of RNA-seq experiments was so good I was seeing red. I’ll be reading everything that Tuuli Lappalainen’s new group at the New York Genome Center writes.
- Hector Corrada Bravo and the crowd at UMD wrote this paper about differential abundance in microbial communities that also made me crazy jealous. Just such a good idea done so well.
- Chad Myers and Curtis Huttenhower continue to absolutely tear it up on networks and microbiome stuff. Just stop guys, you are making the rest of us look bad…
- I don’t want to go to Stanford I want to go to Johns Hopkins.
- Ramnath keeps Ramnathing (def. to build incredible things at a speed that we can’t keep up with by repurposing old tools in the most creative way possible) with rCharts.
- Neo Chung and John Storey invented the jackstraw for testing the association between measured variables and principal components. It is an awesome idea and a descriptive name.
- I wasn’t at Bioc 2013, but I heard from two people who I highly respect and it takes a lot to impress that Levi Waldron gave one of the best talks they’d ever seen. The paper isn’t up yet (I think) but here is the R package with the data he described. His survHd package for fast coxph fits (think rowFtests but with Cox) is also worth checking out.
- John Cook kept cranking out interesting posts, as usual. One of my favorites talks about how one major component of expertise is the ability to quickly find and correct inevitable errors (for example, in code).
- Larry Wasserman’s Simpson’s Paradox post should be required reading. He is shutting down Normal Deviate, which is a huge bummer.
- Andrew Gelman and I don’t always agree on scientific issues, but there is no arguing that he and the stan team have made a pretty impressive piece of software with stan. Richard McElreath also wrote a slick interface that makes fitting a fully Bayesian model match the syntax of lmer.
- Steve Pierson and Ron Wasserstein from ASA are also doing a huge service for our community in tackling the big issues like interfacing statistics to government funding agencies. Steve’s Twitter feed has been a great resource for keeping track of deadlines for competitions, grants, and other deadlines.
- Joshua Katz built these amazing dialect maps that have been all over the news. Shiny Apps are getting to be serious business.
- Speaking of RStudio, they keep rolling out the goodies, my favorite recent addition is interactive debugging.
- I’ll close with David Duvenaud’s HarlMCMC shake:
 ).
). Follow us on twitter
Follow us on twitter 